Chlamydomonas is a single-celled green alga while its close kin Volvox is a multicellular green alga. Both algae are well-studied models and have been sequenced before (see “Why Sequence the Mating Loci of Chlamydomonas reinhardtii and Volvox carteri?”) using Sanger or shotgun sequencing. Researchers hope to get more information from the genomes by re-sequencing the algae using new technologies (see “Why Sequence Chlamydomonas by New Methods?”). In particular, they now want to focus on all the messenger RNAs in the algae to gain a better understanding of how these organisms adapt to changes in their environments.

Chlamydomonas reinhardtii
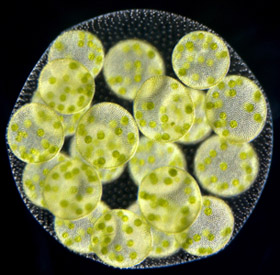
Volvox carteri, Ichiro Nishii, RIKEN
Principal Investigators: Sabeeha Merchant, Univ. of California, Los Angeles
Program: CSP 2009