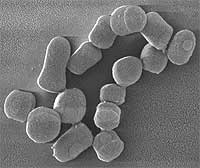
SEM of Arthrobacter chlorophenolicus
Scanning electron micrograph of Arthrobacter chlorophenolicus cells
Bioremediation of toxic pollutants has been extensively studied as a cleanup strategy over the last 40 years, but it is often limited by a lack of microorganisms having the genes and corresponding enzymes necessary to degrade recalcitrant compounds. Another problem is that sometimes the target compound is only partially degraded, leading to accumulation of toxic metabolites. One way to overcome these limitations is to inoculate the contaminated site with cells that have the catabolic enzymes required for complete degradation of those compounds. This is a process referred to as bioaugmentation. One critical challenge of bioaugmentation is the isolation, or creation by genetic modification, of appropriate biodegrading strains.
The microbial world has incredible metabolic diversity that has just begun to be explored. It is commonly estimated that approximately 1% or less of the microbial species on Earth have been isolated and identified. As we continue to isolate and characterize new degrading strains, we might be able to exploit any novel biodegradation potential they may have. As a prime example, actinobacteria are a group of bacteria that are known for their ability to degrade a wide range of toxic pollutants. This group is also known to be particularly well adapted to survival in harsh environments, such as soil. In order to fully utilize the biotechnological potential of actinobacteria for clean-up of polluted environments, it is necessary to first understand basic molecular and physiological aspects affecting the efficacy and performance of the cells in nature. Both candidate strains in this project, Arthrobacter chlorophenolicus and Micrococcus luteus, have novel and interesting environmental applications; for example, bioremediation of recalcitrant chloroaromatic compounds (A. chlorophenolicus), bioremediation of organic pollutants in combination with metal tolerance (M. luteus), and sequestering or bioadsorbtion of metal wastes (M. luteus). Therefore, there are relevant environmental applications for both of these bacteria, representative of many members of the actinobacteria group. In addition, M. luteus may be used for gold concentration from low-abundance ores. The genome sequence information should provide us novel information about the genes responsible for these environmentally interesting properties in actinobacteria and how to take advantage of these properties for environmental applications.
Principal Investigator: Janet Jansson (Swedish Univ. of Agricultural Sciences) and Charles Greenblatt (Hebrew Univ.)