Researchers have devised a novel method to exploit relationships between bacteria and archaea for a new set of gene markers
The Science
The researchers developed a new way to identify gene markers in bacteria and the primitive microorganisms classified in the kingdom known as Archaea. Dubbed, PhyEco (for phylogenetic and phylogenetic ecology) this strategy can be used to identify organisms in metagenome studies (collected from a environmental microbial community, not just colonies of a single type of organism), including those that have not been cultured in the lab.
The Impact
New tools are needed to efficiently and accurately analyze rapidly accumulating genome and metagenome data. By identifying a new set of gene markers for bacteria and archaea, researchers seek to extend this strategy to an automated method to enhance the library of bacterial and archaeal gene markers available to the research community. In particular, the PhyEco marker identification effort would benefit the phylogeny-driven initiative at the DOE Joint Genome Institute, called the Genomic Encyclopedia of Bacteria and Archaea (GEBA) which focuses on cultured organisms and culture-independent genome sampling efforts.
Summary
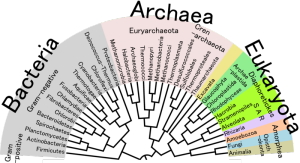
Gene markers that highlight relationships between organisms can also be used to study microbes in their communities. Image via Wikimedia Commons.
Researchers studying the evolutionary relationships between species of bacteria and archaea have relied on a handful of genetic “markers” that are conserved over most species (e.g., small subunit rRNA), but have enough genetic diversity that it can track evolutionary changes over time. These methods, however, have their limitations, including the introduction of bias — favoring some taxonomic groups over others. As researchers studying bacteria have moved away from single lab-cultured microorganisms and toward metagenomic studies that can include hundreds of bacterial species, gene markers have been used more and more to study diversity in microbial communities. Gene marker sequences are used to measure the abundance of bacterial and archaeal species in a habitat and establish an organism’s position on a phylogenetic tree. But trees built from one gene do not always reflect the true evolution history.
Increasing the number of gene markers would alleviate that problem, but the task of developing new markers has typically been onerous. To address this issue, the authors of this paper have developed an automated technique for quickly evaluating potential gene markers from among a large library of genes, described in a paper published October 17 in the journal, PLoS ONE.
The researchers found 40 markers that can be used for all bacteria and archaea, another 114 that can be used just for bacteria and hundreds to thousands when they studied smaller subgroupings of related bacteria. Many of these are associated with translating DNA to RNA (the molecules that deliver the genetic message from the DNA to other parts of the cell). The researchers have also shared the gene family data and computer code for evaluating them online.
Contact
Jonathan Eisen
UC Davis
[email protected]
Funding
The Gordon and Betty Moore Foundation; the DOE Office of Science, Office of Biological and Environmental Research.
Publication
Wu D, Jospin G, Eisen JA (2013) Systematic Identification of Gene Families for Use as ‘‘Markers’’ for Phylogenetic and Phylogeny-Driven Ecological Studies of Bacteria and Archaea and Their Major Subgroups. PLoS ONE 8(10): e77033.
doi:10.1371/journal.pone.0077033
Related Links
http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0077033