We’re almost ready to read the bases by capillary electrophoresis. But first the reaction must be cleaned up. To remove the unused product of the sequencing reaction,
![]() Beads (magnetic microparticles) are added to the sequencing reaction solution. DNA, but not the excess dye terminators, is induced to attach to the beads.
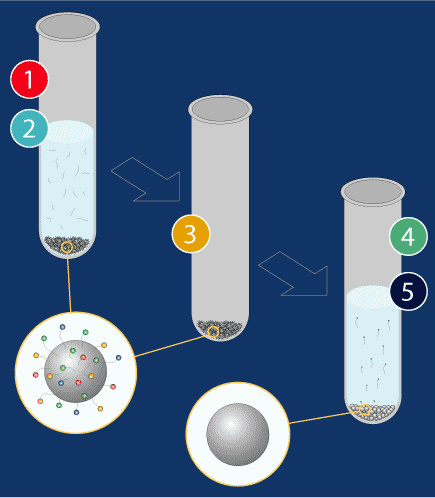
Beads (magnetic microparticles) are added to the sequencing reaction solution. DNA, but not the excess dye terminators, is induced to attach to the beads.
![]() Magnets draw down the DNA and beads so the waste solution can be aspirated.
Magnets draw down the DNA and beads so the waste solution can be aspirated.
![]() The beads are washed with ethanol and then dried.
The beads are washed with ethanol and then dried.
![]() Water is then added to the beads, and the DNA strands detach from the beads and reenter solution because they have higher affinity for water than for the beads.
Water is then added to the beads, and the DNA strands detach from the beads and reenter solution because they have higher affinity for water than for the beads.
![]() Magnets are then used to secure the beads while the pipettor removes the clean DNA/water solution to a new plate.
Magnets are then used to secure the beads while the pipettor removes the clean DNA/water solution to a new plate.