Ten µl from each well is transferred to a new 384-well PCR plate to be loaded onto a MegaBACE 4000 or ABI 3730xl sequencer. Each plate is tracked with a barcode.

Bar codes are used to track plates to be sequenced.
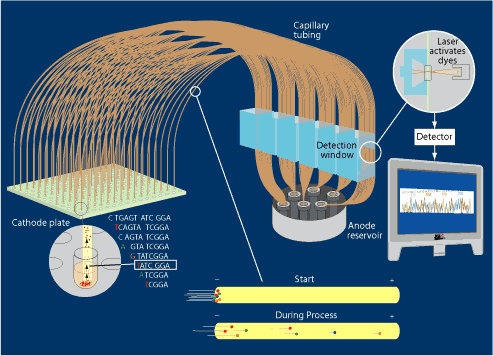
Our wells now contain billions of copies of our original fragment, but they are all different lengths and each ends with a dye terminator. The fragments, which have a negative charge, move through the capillaries of the sequencing machine toward the positively charged pole. Shorter fragments move faster than longer ones. At the detection window, a laser excites the dyes. A detector “reads” the colors, one at a time, to determine the sequence of the tagged As, Ts, Gs, and Cs.
Results are shown as an electropherogram showing a peak for each base. From the peak heights and widths, a Phred score is assigned to each individual base. A high Phred score indicates a high certainty as to the identity of that particular base.
Each sequencing machine can run 384 samples (lanes) through its 384 capillary tubes at one time.