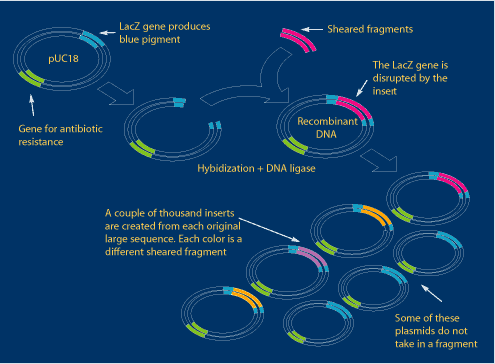
All the small random fragments generated by shearing are inserted, or “ligated,” into a plasmid (a loop of nonessential bacterial DNA) called pUC18. The pUC18 vector allows for propagation and replication in Escherichia coli bacteria (E. coli). Each plasmid has a gene for antibiotic resistance (green in the diagram below) as well as a LacZ gene (blue) that produces blue pigment. These two genes will be important later. The fragments, shown here in various colors, are inserted in the middle of the LacZ gene.