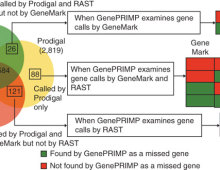
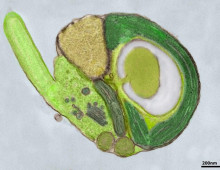
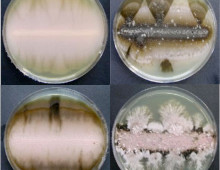
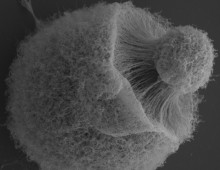
Noteworthy DOE JGI papers in Nature Methods
Three publications are featured in the Special 10th Anniversary issue. The Science: To mark its 10th anniversary, the journal Nature Methods released a Special Issue highlighting impactful articles. Among these noteworthy articles are three from DOE JGI researchers. The Impact: Microbial Sequencing is one of the areas of methods development highlighted by the journal, and… [Read More]