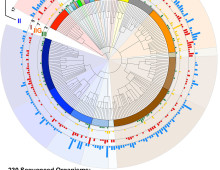
Modeling Microbial Networks in an Oxygen Minimum Zone
UBC team develops predictive marine microbiome math model. The Science With help from two DOE national user facilities, a team at the University of British Columbia (UBC) has developed a math model that could help researchers and policy makers track the impact of climate change on the microbial networks that drive the world’s marine ecosystems…. [Read More]