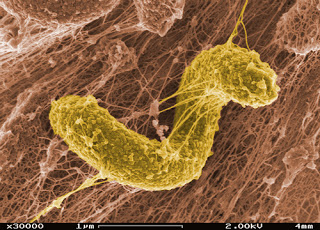
Color-enhanced photo from SEM image of sulfate-reducing Desulfovibriodesulfuricansin a biofilm matrix. (Image courtesy of PNNL via Flickr/Creative Commons)
Mercury pollution in aquatic environments has been a concern, though the contaminants are mainly sourced from industrial processes and fossil fuel combustion. Isolated from Chesapeake Bay sediment, the sulfate-reducing bacterium, Desulfovibriodesulfuricansstrain ND132, can also produce the human neurotoxin methylmercury.
“What is not known are the genes or the proteins that allow these bacteria to mediate the transformation,” said Oak Ridge National Laboratory’s Steven Brown.
To help answer the question and understand how a bacterium can convert inorganic mercury into a more toxic compound, Brown and colleagues at ORNL collaborated with DOE JGI scientists including DOE JGI Microbial Program head Tanja Woyke and at ORNL and Los Alamos National Laboratory to sequence the genome of D. desulfuricansND132. With the genomic information, researchers hope to understand how to manage the impacts of mercury pollution.
In the April 2011 issue of the Journal of Bacteriology, the team reported that the 3.8-million base genome was sequenced using 454 and Illumina platforms, and candidate protein-encoding gene models were predicted using Prodigal and GenePRIMP software developed at the DOE JGI. The genome is currently described as noncontiguous finished (there are six gaps remaining), indicating a high-quality assembly with automated and manual improvements.