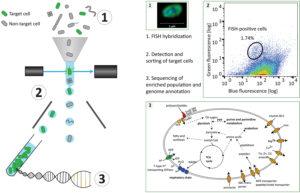
In the protocol developed by Bernhard Fuchs’s team, bacterial groups are enriched in three steps. 1. Specific bacterial cells are fluorescently labeled by fluorescence in situ hybridization (FISH). 2. These targeted cells are enriched based on their FISH signal via flow cytometric cell sorting. 3. The enriched population is then sequenced and further analyzed. Genome annotation reveals putative functional repertoires. (Overview and Figure 1: Courtesy of Anissa Grieb, Figures 2 and 3: Grieb A et al. A pipeline for targeted metagenomics of environmental bacteria. Microbiome. 8:21;2020 Feb 17, Figures 4 and 6, http://creativecommons.org/licenses/by/4.0/)