A deep dive into microbial genomics reveals one bacterial species is made of four ecologically distinct groups with different lifestyles.

Boeuf and colleagues collected samples of SAR324 microbial communities from this research vessel, the Kilo Moana. (School of Ocean And Earth Science And Technology at University of Hawaii at Manoa)
The Science
The bacterium SAR324 is unusually cosmopolitan. In the ocean’s North Pacific Subtropical Gyre, microbes tend to stay localized at different depths. But SAR324 can be found throughout the water column, from the warm, well-lit surface, to the blue-lit twilight zone, to the continuous pressure of the dark abyss, 4000 m (2.5 miles) deep. Scientists have wondered, how can SAR324 exist in so many varied environments? Now, a recent study published in Microbiome has uncovered that SAR324 encompasses four subgroups, adapted to different oceanic depths and relying on different ways of living.
The Impact
Microbes help regulate the global carbon cycle — fixing, transforming, and respiring carbon. Understanding the metabolisms that SAR324 and other microbes use could inform scientists’ understanding of carbon fluxes in the environment. Moreover, the deep ocean is the largest inorganic reservoir of carbon on earth; because SAR324 — one of the most abundant organisms in the deep ocean — can fix carbon, SAR324 is able to influence this carbon pool.
Summary
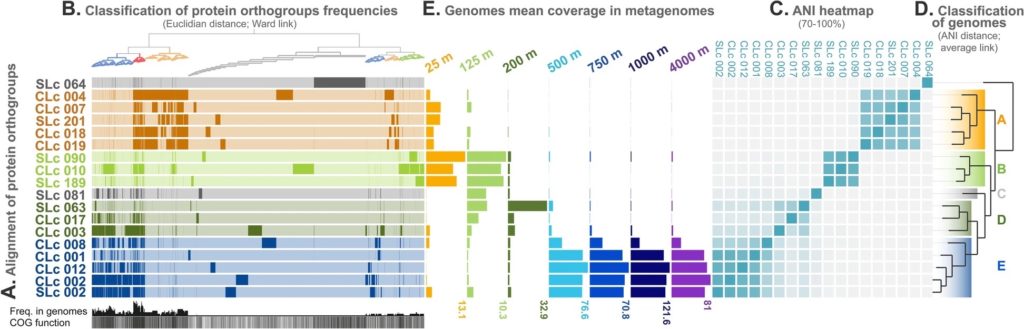
An excerpt from Figure 1 of the study. The data show that SAR324 from different depths cluster into their own groups, or ecotypes. See the paper for more. (Boeuf, D. et al. Metapangenomics reveals depth-dependent shifts in metabolic potential for the ubiquitous marine bacterial SAR324 lineage. Microbiome. 2021 Aug 13. https://doi.org/10.1186/s40168-021-01119-5)
To study the lifestyles of SAR324, lead author Dominique Boeuf and colleagues set sail on research cruises in 2015 and 2016. At Station ALOHA, site of the 30-years-long Hawaii Ocean Time-series (HOT), they collected samples of the ocean’s microbial communities periodically at different depths. In order to determine the genome structure of SAR324 cells and their gene expression, the team then sequenced community genomes and expressed gene transcripts (metagenomes, metatranscriptomes), as well as the genomes of single cells of the uncultivated SAR324. The constellation of data gave the team unprecedented resolution to study the network of genes that contribute to the metabolisms that individual SAR324 cells can carry out. The work on single cells was done in collaboration with scientists Tanja Woyke and Rex Malmstrom at the U.S. Department of Energy Joint Genome Institute (JGI), a DOE Office of Science User Facility located at Lawrence Berkeley National Laboratory (Berkeley Lab), through the JGI Community Science Program. The metagenomes were also processed using the JGI-developed software suite, BBTools, and relying on the JGI Integrated Microbial Genomes & Microbiomes (IMG/M) system.
By analyzing the metagenomes, metatranscriptomes and single cell genomes, Boeuf — then a postdoctoral associate in Ed DeLong’s group at the University of Hawaii at Manoa — and colleagues were able to see that SAR324 was made up of four ecologically distinct clusters, or ecotypes. These ecotypes fall into two main groups: two ecotypes dwell in the sunlit upper waters (above 200 m, or 656 ft), and have genes to support a photoheterotrophic lifestyle. In other words, they snack on fixed carbon if they can get it, rather than make it themselves, but use sunlight to help in the struggle to survive near the surface. The other two ecotypes, on the other hand, thrive in the dark, below 200 m (656 ft). They have genes supporting a chemolithoautotrophic way of life; they can fix carbon dioxide, but they drive that high-energy reaction using simple inorganic sulfur-containing chemicals instead of light.
Boeuf pointed out that, without the JGI, the discoveries made in this project wouldn’t have been possible. Scientists hadn’t known that SAR324 encompassed these different ecotypes because, historically, they have used the sequence of a “marker gene,” namely the 16S ribosomal RNA (rRNA) gene, to mark or identify microbial species. All SAR324 have very closely related 16S rRNA genes, but these genes mask the striking diversity of their genomic capabilities. According to Boeuf, 16S rRNA genes are like stamps on packages: Focus only on the stamps, and you might miss the rich contents they’re attached to.
Contacts:
BER Contact
Ramana Madupu, Ph.D.
Program Manager
Biological Systems Sciences Division
Office of Biological and Environmental Research
Office of Science
US Department of Energy
[email protected]
PI Contact
Edward F. DeLong
University of Hawaii at Manoa
[email protected]
Funding:
This research was supported by grants from the Simons Foundation (#329108 and #721223 to EFD) and Gordon and Betty Moore Foundation grant GBMF #3777 (to EFD). The work conducted by the US Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported under contract number DE-AC02-05CH11231. This work is a contribution of the Simons Collaboration on Ocean Processes and Ecology and the Daniel K. Inouye Center for Microbial Oceanography: Research and Education.
Publication:
- Boeuf et al. Metapangenomics reveals depth-dependent shifts in metabolic potential for the ubiquitous marine bacterial SAR324 lineage. 2021 August 13. doi: 10.1186/s40168-021-01119-5
Related Links:
- Video abstract by Researchsquare: “Depth-related metabolic diversity in the marine bacterial clade SAR324“
- Ed DeLong at the 2015 DOE JGI Genomics of Energy & Environment Meeting
- JGI Single Cell Genomics Group
- Engagement Webinar: How to Successfully Apply for a CSP Proposal
Byline: Alison F. Takemura