Phil Hugenholtz on metagenomics and ecogenomics in Australian Life Scientist
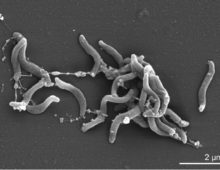
In research published in Nature in 2007, Hugenholtz, along with collaborators from the California Institute of Technology and Diversa (now Verenium) Corporation, used metagenomics to detail the process by which a dry wood feeding termite, a Nasutitermes species, breaks down cellulose. They generated 62 million base pairs – a “drop in the ocean by today’s… [Read More]