The rhizosphere is the space in, on and around the plant roots where microbes in the plant interact with the microbes in the soil. These interactions can be critical in determining plant health and yield, information that could be useful for developing bioenergy feedstocks.
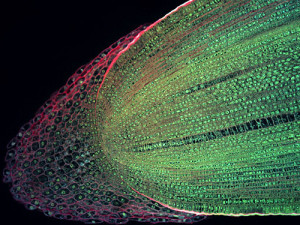
Longitudinal section of a root tip of Maize (Zeamays). To the left of the image,
the large, loosely packed cells of the root cap can be seen. These cells protect the actively dividing undifferentiated plant tissue as the root grows down through the soil.
Creative Commons Attribution-Noncommercial-Share Alike 2.0 Generic License by Science and Plants for Schools
To learn more about these influences, the DOE Joint Genome Institute initiated a Rhizosphere Grand Challenge project involving the model plant Arabidopsis. A similar project was also initiated with maize.
In the study that appeared April 16, 2013 in Proceedings of the National Academy of Sciences, DOE JGI researchers were part of a team that characterized the rhizosphere bacterial diversity of maize plants in order to identify genes involved in plant-microbe interactions and how the environments might influence such interactions.
The team sequenced and analyzed rhizosphere microbial DNA taken from soil samples of maize fields collected in two climatic regions: Illinois and Missouri; and northeastern New York. They noted in their findings that despite the geography, the microbes from the Midwestern fields were less similar with each other; the Missouri soil microbes had greater similarity with the New York microbes. Other findings suggest that the microbial community around maize is strongly influenced by a few key genes rather than several alleles in concert.