Differences between genome sequences warrant fungal reclassifications.
The Science:
Researchers sequenced four samples of Aureobasidium fungi from various regions of the world to learn more about their potential biotechnological applications.
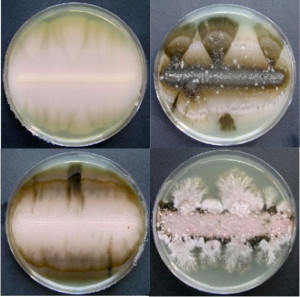
Aureobasidium species sequenced, clockwise from top left: A. pullulans; A. namibiae; A. subglaciale; and, A. melogenum. (Cene Gostincar, University of Ljubljana)
The Impact:
Comparative genomic analyses reveal genes that make the fungi capable of adapting to a wide range of environments and stresses, which can be useful in a variety of industrial applications from improving biofuels production to biomaterials and in medicine.
Summary
Grown on a Petri dish, the yeast Aureobasidium pullulans appears innocuous, and yet this fungus has been found from the tropics to the poles. Additionally, the fungus has been used as a biocontrol agent for plants, is a known producer of a biomaterial that could be used for packaging food and drugs, and is considered one of the most effective fungi at breaking down xylan in plant cell walls.
For these reasons, in 2011, researchers led by Nina Gunde-Cimerman at the University of Ljubljana, Slovenia proposed that the genome of A. pullulans be sequenced under the U.S. Department of Energy Joint Genome Institute’s (DOE JGI) Community Science Program. Under the CSP proposal, four varieties of A. pullulans were sequenced using samples collected from: hypersaline water in a Slovenian saltern; dolomitic marble in the Namibian desert; subglacial ice in Norway; and, a public fountain in Thailand. As reported in a paper published ahead online July 1, 2014 in BMC Genomics, researchers from the DOE JGI and the University of Slovenia found that the differences found in the genomes of the four varieties sequenced led them to propose that these fungi be considered distinct Aureobasidium species: A. pullulans; A. namibiae; A. subglaciale; and, A. melogenum.
One of the team’s findings is that the enzyme families involved in breaking down plant material are “significantly enriched” in A. pullulans from the hypersaline environment compared to the other three species sequenced, while the same enzyme families are reduced in A. melogenum from the Thai fountain. The team also reported finding increased concentrations of compounds that protect against a variety of stressors, providing these fungi with the ability to tolerate a wide range of environments.
“The investigated genomes of these four Aureobasidium species contain an abundance of different families of extracellular enzymes, which are important for the degradation of the plant cell-wall material,” the team reported. “Their diversity is comparable to the phylogenetically related plant pathogens. The differences in the abundance of the extracellular enzymes and also in the higher numbers of sugar transporters reflect the ecological preferences of the four studied Aureobasidium species, especially for the plant-associated lifestyle of A. pullulans….The genome sequences of the described four Aureobasidium species are expected to facilitate the exploitation of the substantial biotechnological potential of these fungi.”
Contact
Igor Grigoriev
DOE Joint Genome Institute
[email protected]
Funding
- U.S. Department of Energy Office of Science
- Slovenian Research Agency
- European Regional Development Fund
- Slovenian Ministry of Higher Education, Science and Technology
Publication
Gostinçar C et al. Genome sequencing of four Aureobasidium pullulans varieties: biotechnological potential, stress tolerance, and description of new species. BMC Genomics. 2014 Jul 1;15(1):549. [Epub ahead of print] http://www.biomedcentral.com/1471-2164/15/549