The genus Desulfurococcus represents a unique clade (a group of organisms all descended from a common ancestor) in the domain of archaea that is not currently represented in the publicly available genomic databases. This scenario will change to some extent as Desulfurococcus mucosus genome sequence is determined under the GEBA (Genomic Encyclopedia for Bacteria and Archaea) Pilot Project of the JGI.
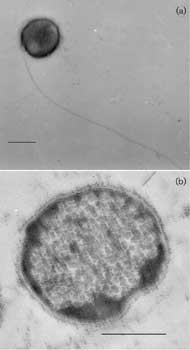
Electron micrographs of desulfurococcus cells of strain Z-1312T. (a) Negatively stained whole cell showing a single flagellum. (b) Thin section. Bars, 0.5 µm. Image from Perevalova, A. A., et al., Int J Syst Evol Microbiol 55:995-999, 2005, reproduced by permission.
The motivation for this project comes from the observation that Desulfurococcus fermentans is the only known archaeon that breaks down cellulose, and that, unlike most known microorganisms that carry out fermentation, it produces hydrogen in the presence of hydrogen while fermenting cellulose and starch without experiencing an inhibition of growth. On the other hand, the starch-utilizing Desulfurococcus amylolyticus and starch non-utilizing and solely peptide-fermenting Desulfurococcus mobilis and D. mucosus are inhibited by hydrogen and stimulated by sulfur; D. fermentans is not stimulated by sulfur. Since these organisms are closely related, differences in their genomes will highlight differences in their metabolisms.
A comparative genomics investigation involving D. fermentans, D. amylolyticus, D. mobilis, and D. mucosus will allow a rapid development of hypotheses about the special molecular tools and regulatory processes that D. fermentans and D. amylolyticus utilize for degrading starch and those that D. fermentans employs for fermenting cellulose and producing hydrogen. It will help to reveal the finer details that distinguish proton reduction (producing hydrogen) from sulfur reduction in fermentative archaea and provide hypotheses concerning the evolution of these two fermentation pathways. In addition, since a strict dependence on sulfur is found in several hyperthermophilic fermentative archaea, the genome data will help to define the evolutionary and metabolic relationships of the Desulfurococcus species with their archaeal relatives.
Principal Investigators: Biswarup Mukhopadhyay (Virginia Bioinformatics Inst., Virginia Polytechnic Inst. and State Univ.)
Program: CSP 2009