The oceans cover more than two-thirds of the planet’s surface, and absorb a significant portion of the carbon dioxide in the atmosphere. Keeping the carbon trapped in the oceans depends on two main factors: the rate of particle production and the rate of particle dissolution. One of the challenges in understanding the role the marine microbial communities play in the global carbon cycle is having genomic information for reference. Though many microbial genomes have been sequenced, not all of them represent the highly abundant organisms in the marine environment.
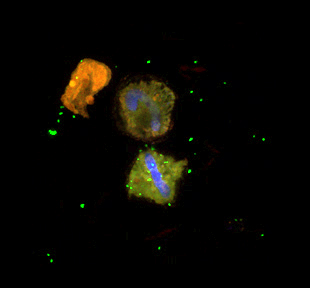
Photo: Confocal image of field populations of RCA cluster bacteria (identical 16S sequence as strain LE17) collected during a bloom of the dinoflagellate Lyngulodinium polyedrum (red = chlorophyll autofluorescence of the dinoflagellates; blue = DNA from the dinoflagellate, green = RCA cluster specific CARD-FISH probe). Strain LE17 attaches to and kills the dinoflagellate cells in culture. Image by Xavier Mayali, LLNL
Researchers working on understanding the roles that marine microbial communities play in the global carbon cycle can reference genomic information from two highly abundant organisms – Prochlorococcus and Pelagibacter (aka SAR11) – that have been sequenced. This project focuses on sequencing a third, highly-abundant marine bacterial group. Having a third such set of genomic data allows researchers to better understand and ultimately predict ecosystem responses to environmental stressors such as climate change and ocean acidification.
Principal Investigators: Xavier Mayali, Lawrence Livermore National Laboratory
Program: CSP 2010