The poet John Donne once noted that no man is an island, and the same can be argued for bacteria. Dehalococcoides ethenogenes bacteria are often found in a community of other microorganisms at groundwater sites contaminated with compounds such as tetrachloroethne and trichloroethene. These chemicals are among most the pervasive organic groundwater pollutants because they’re often used by industry as cleansers or degreasers. Of all the Dehalococcoides strains identified thus far, D. ethenogenes is the only bacterial strain known to be able to break down the chlorinated groundwater pollutants, and the only strain that has been sequenced. Part of the reason for the latter is the size of the bacteria’s genome, which is small enough that it can be easily separated from the rest of the microbial community for study. But when cultivated apart from the microbial community, D. ethenogenes needs to be supplied with nutrients to thrive.
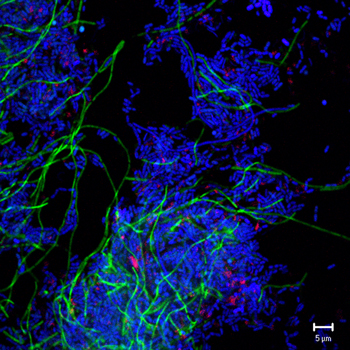
Photo: Fluorescence in situ hybridization image of Dehalococcoides cells (pink), filamentous microbes affiliated to Archaea (green), and other not yet characterized microbes (blue) with unknown function in a dechlorinating methanogenic culture, courtesy of Natuschka M. Lee (Techn. University of Munich, Germany) and Frank E. Löffler (Georgia Institute of Technology, USA)
Now researchers want to learn more about these other microbes found with D. ethenogenes to understand the interrelationships within the community, from the nutrients they might supply each other to the roles they might play in breaking down chlorinated pollutants. Additionally, the community being sequenced also contains microbes known to be of interest to DOE not just for bioremediation purposes but for bioenergy as well. One microbe is a hydrogen producer while two others are methanogenic.
Principal Investigators: Ruth Richardson (Cornell University)
Program: CSP 2009