Heterotrophic nanoflagellates are a group of marine microbes that prey on other microbes, such as bacteria and phytoplankton. Bacteria and phytoplankton constitute a dominant fraction of the living biomass in marine ecosystems. Their fates are dictated largely by two major forces: predation by protists like the nanoflagellates, and cell death induced by viruses. These two forces have very different consequences on the fate of phytoplankton and bacterial carbon, affecting whether it can be sequestered into larger sinking particles/cells (via predation) or simply released back into the environment (viruses). Thus predatory protists play critical roles in marine carbon cycling.
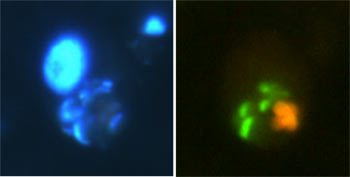
Marine heterotrophic protist with prey cells in its gut. The orange comes from several Synechococcus (a photosynthetic marine cyanobacterium) cells and the green from greeen fluorescent protein-labelled Vibro cholerae cells (the bacterium that can cause cholera). The large blue region in the left panel is the nucleus of the eukaryotic predator (protist). Photos: AZ Worden.
However, little is known about the basic biology of predatory nanoflagellates. While rates of consumption, growth, and even assimilation and egestion (discharge of undigested food), have been quantified, the underlying mechanisms remain largely unexplored. Thus the molecular underpinnings of heterotrophic nanoflagellate behavior and interactions with prey are still unknown. Currently no sequenced genomes are available from these organisms.
Thus, this project is designed to allow researchers to “listen to the message” generated by these organisms and to deduce real-time behavior based on genomic composition and mRNA expression. An international group of investigators led by Alexandra Worden (Monterey Bay Aquarium Research Institute) will collaborate in sequencing the genomes of three target species. These are Paraphysomonas imperforata, a widespread marine heterotroph; Ochromonas a marine mixotroph (mixotrophs can synthesize nutrients from both inorganic and organic compounds, whereas heterotrophs must rely on organic material alone); and a third organism, Spumella, which is a heterotrophic nanoflagellate common in freshwater systems. This suite of organisms provides a unique opportunity for comparative studies (e.g. heterotrophy vs. phototrophy and mixotrophy, marine vs. freshwater habitats) that will greatly facilitate carbon cycling research, addressing evolutionary developments, novel biochemical pathways, and the integration of potentially competing metabolic processes.
Principal Investigators: Andrew Allen (J.Craig Venter Inst.), Jens Boenigk (Austrian Acad. of Sciences), David A. Caron and Karla B. Heidelberg (Univ. of S. California), Emily Roberts (Swansea Univ.), and Alexandra Z. Worden (Monterey Bay Aquarium Res. Inst.).
Program: CSP 2009