Trichoderma virens is a haploid, filamentous hyphomycete (a subclass of fungi). As strains of T. virens are used to protect many crops from a variety of pathogens, this species is a model system for elucidating the mechanisms of biological control. Mechanisms being investigated include mycoparasitism and antibiosis (direct interaction with the pathogen), induction of host plant resistance, metabolism of pathogen germination stimulants released by seeds, and increased tolerance to stress by enhancing plant growth (indirect effects). Since this fungus is present in most soils throughout the world, isolates affect ecosystem health and productivity. This occurs both through interactions with pathogens and through induced changes in plant chemistry, which influence growth and interactions with insect pests.
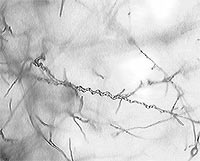
Coiling of Trichoderma hyphae around Rhizoctonia (a basidiomycete pathogen). The coiling response of T. virens is induced by the presence of host hyphae and precedes penetration of the host.
Interestingly, interactions with plants develop as ingress into the root epidermal cells is initiated by hyphae of T. virens, but further branching into other root cells does not occur. The ability to rapidly and extensively colonize roots, induce resistance mechanisms in plants, and enhance the growth of plants while remaining an aggressive parasite of plant pathogenic fungi places T. virens in a very unique biological niche. Stated simply, T. virens possesses combinations of traits within a single organism that are usually associated with distinct species of symbionts or pathogens. In addition to the production of novel secondary metabolites (of interest to the pharmaceutical and agricultural sectors), T. virens has the capacity to degrade hazardous compounds, including pesticides, polyphenols, and polyaromatic hydrocarbons, and to sequester heavy metals. Whether as a bioaugmentation agent or as part of a phytoremediation system, T. virens possesses metabolic diversity that exceeds that of most of the extant sequenced fungal genomes. The genome sequence and subsequent functional and comparative genetic analyses will provide a context for understanding the enigmatic biology of this fungus.
CSP project participants: Charles M. Kenerley (proposer), Daniel J. Ebbole, Heather H. Wilkinson, and Michael Thon (Texas A&M Univ.); and Alfredo Herrera-Estrella (Center for Research and Advanced Studies, Mexico).