Single species reveals a myriad of genomic variations
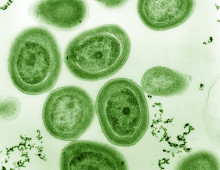
Single cell genomics reveals diversity of cyanobacteria subpopulations. The Science: Single cell genomics allowed researchers to examine the subpopulations of the marine microbe Prochlorococcus, affording a glimpse at “a new dimension of microdiversity.” The Impact: The cyanobacterial subpopulations were found to be distinct, comprised of “genomic backbones” made of highly conserved core gene alleles and… [Read More]